matlab2021a
这里,采用的pointnet网络结构如下图所示:
在整体网络结构中,
首先进行set abstraction,这一部分主要即对点云中的点进行局部划分,提取整体特征,如图可见,在set abstraction中,主要有Sampling layer、Grouping layer、以及PointNet layer三层构成,sampling layer即完成提取中心点工作,采用fps算法,而在grouping中,即完成group操作,采用mrg或msg方法,最后对于提取出得点,使用pointnet进行特征提取。在msg中,第一层set abstraction取中心点512个,半径分别为0.1、0.2、0.4,每个圈内的最大点数为16,32,128。
Sampling layer
采样层在输入点云中选择一系列点,由此定义出局部区域的中心。采样算法使用迭代最远点采样方法 iterative farthest point sampling(FPS)。先随机选择一个点,然后再选择离这个点最远的点作为起点,再继续迭代,直到选出需要的个数为止相比随机采样,能更完整得通过区域中心点采样到全局点云
Grouping layer
目的是要构建局部区域,进而提取特征。思想就是利用临近点,并且论文中使用的是neighborhood ball,而不是KNN,是因为可以保证有一个fixed region scale,主要的指标还是距离distance。
Pointnet layer
在如何对点云进行局部特征提取的问题上,利用原有的Pointnet就可以很好的提取点云的特征,由此在Pointnet++中,原先的Pointnet网络就成为了Pointnet++网络中的子网络,层级迭代提取特征。
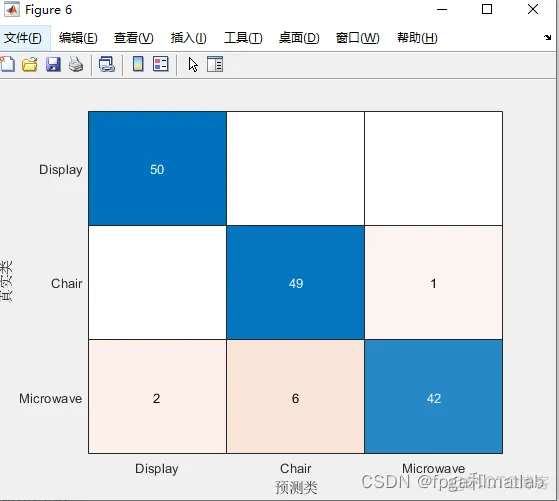
clc;clear;close all;warning off;addpath(genpath(pwd));rng('default')%****************************************************************************%更多关于matlab和fpga的搜索“fpga和matlab”的CSDN博客:%matlab/FPGA项目开发合作%https://blog.csdn.net/ccsss22?type=blog%****************************************************************************dsTrain = PtCloudClassificationDatastore('train');dsVal = PtCloudClassificationDatastore('test');ptCloud = pcread('Chair.ply');label = 'Chair';figure;pcshow(ptCloud)xlabel("X");ylabel("Y");zlabel("Z");title(label)dsLabelCounts = transform(dsTrain,@(data){data{2} data{1}.Count});labelCounts = readall(dsLabelCounts);labels = vertcat(labelCounts{:,1});counts = vertcat(labelCounts{:,2});figure;histogram(labels);title('class distribution')rng(0)[G,classes] = findgroups(labels);numObservations = splitapply(@numel,labels,G);desiredNumObservationsPerClass = max(numObservations);filesOverSample=[];for i=1:numel(classes)if i==1 targetFiles = {dsTrain.Files{1:numObservations(i)}};else targetFiles = {dsTrain.Files{numObservations(i-1)+1:sum(numObservations(1:i))}};end% Randomly replicate the point clouds belonging to the infrequent classesfiles = randReplicateFiles(targetFiles,desiredNumObservationsPerClass);filesOverSample = vertcat(filesOverSample,files');enddsTrain.Files=filesOverSample; dsTrain.Files = dsTrain.Files(randperm(length(dsTrain.Files)));dsTrain.MiniBatchSize = 32;dsVal.MiniBatchSize = dsTrain.MiniBatchSize;dsTrain = transform(dsTrain,@augmentPointCloud);data = preview(dsTrain);ptCloud = data{1,1};label = data{1,2};figure;pcshow(ptCloud.Location,[0 0 1],"MarkerSize",40,"VerticalAxisDir","down")xlabel("X");ylabel("Y");zlabel("Z");title(label)minPointCount = splitapply(@min,counts,G);maxPointCount = splitapply(@max,counts,G);meanPointCount = splitapply(@(x)round(mean(x)),counts,G);stats = table(classes,numObservations,minPointCount,maxPointCount,meanPointCount)numPoints = 1000;dsTrain = transform(dsTrain,@(data)selectPoints(data,numPoints));dsVal = transform(dsVal,@(data)selectPoints(data,numPoints));dsTrain = transform(dsTrain,@preprocessPointCloud);dsVal = transform(dsVal,@preprocessPointCloud);data = preview(dsTrain);figure;pcshow(data{1,1},[0 0 1],"MarkerSize",40,"VerticalAxisDir","down");xlabel("X");ylabel("Y");zlabel("Z");title(data{1,2})inputChannelSize = 3;hiddenChannelSize1 = [64,128];hiddenChannelSize2 = 256;[parameters.InputTransform, state.InputTransform] = initializeTransform(inputChannelSize,hiddenChannelSize1,hiddenChannelSize2);inputChannelSize = 3;hiddenChannelSize = [64 64];[parameters.SharedMLP1,state.SharedMLP1] = initializeSharedMLP(inputChannelSize,hiddenChannelSize);inputChannelSize = 64;hiddenChannelSize1 = [64,128];hiddenChannelSize2 = 256;[parameters.FeatureTransform, state.FeatureTransform] = initializeTransform(inputChannelSize,hiddenChannelSize,hiddenChannelSize2);inputChannelSize = 64;hiddenChannelSize = 64;[parameters.SharedMLP2,state.SharedMLP2] = initializeSharedMLP(inputChannelSize,hiddenChannelSize);inputChannelSize = 64;hiddenChannelSize = [512,256];numClasses = numel(classes);[parameters.ClassificationMLP, state.ClassificationMLP] = initializeClassificationMLP(inputChannelSize,hiddenChannelSize,numClasses);numEpochs = 60;learnRate = 0.001;l2Regularization = 0.1;learnRateDropPeriod = 15;learnRateDropFactor = 0.5;gradientDecayFactor = 0.9;squaredGradientDecayFactor = 0.999;avgGradients = [];avgSquaredGradients = [];[lossPlotter, trainAccPlotter,valAccPlotter] = initializeTrainingProgressPlot;% Number of classesnumClasses = numel(classes);% Initialize the iterationsiteration = 0;% To calculate the time for trainingstart = tic;% Loop over the epochsfor epoch = 1:numEpochs % Reset training and validation datastores. reset(dsTrain); reset(dsVal); % Iterate through data set. while hasdata(dsTrain) % if no data to read, exit the loop to start the next epoch iteration = iteration + 1; % Read data. data = read(dsTrain); % Create batch. [XTrain,YTrain] = batchData(data,classes); % Evaluate the model gradients and loss using dlfeval and the % modelGradients function. [gradients, loss, state, acc] = dlfeval(@modelGradients,XTrain,YTrain,parameters,state); % L2 regularization. gradients = dlupdate(@(g,p) g + l2Regularization*p,gradients,parameters); % Update the network parameters using the Adam optimizer. [parameters, avgGradients, avgSquaredGradients] = adamupdate(parameters, gradients, ... avgGradients, avgSquaredGradients, iteration,learnRate,gradientDecayFactor, squaredGradientDecayFactor); % Update the training progress. D = duration(0,0,toc(start),"Format","hh:mm:ss"); title(lossPlotter.Parent,"Epoch: " + epoch + ", Elapsed: " + string(D)) addpoints(lossPlotter,iteration,double(gather(extractdata(loss)))) addpoints(trainAccPlotter,iteration,acc); drawnow end % Create confusion matrix cmat = sparse(numClasses,numClasses); % Classify the validation data to monitor the tranining process while hasdata(dsVal) data = read(dsVal); % Get the next batch of data. [XVal,YVal] = batchData(data,classes);% Create batch. % Compute label predictions. isTrainingVal = 0; %Set at zero for validation data YPred = pointnetClassifier(XVal,parameters,state,isTrainingVal); % Choose prediction with highest score as the class label for % XTest. [~,YValLabel] = max(YVal,[],1); [~,YPredLabel] = max(YPred,[],1); cmat = aggreateConfusionMetric(cmat,YValLabel,YPredLabel);% Update the confusion matrix end % Update training progress plot with average classification accuracy. acc = sum(diag(cmat))./sum(cmat,"all"); addpoints(valAccPlotter,iteration,acc); % Update the learning rate if mod(epoch,learnRateDropPeriod) == 0 learnRate = learnRate * learnRateDropFactor; end reset(dsTrain); % Reset the training data since all the training data were already read % Shuffle the data at every epoch dsTrain.UnderlyingDatastore.Files = dsTrain.UnderlyingDatastore.Files(randperm(length(dsTrain.UnderlyingDatastore.Files))); reset(dsVal);endcmat = sparse(numClasses,numClasses); % Prepare sparse-double variable to do like zeros(2,2)reset(dsVal); % Reset the validation datadata = readall(dsVal); % Read all validation data[XVal,YVal] = batchData(data,classes); % Create batch.% Classify the validation data using the helper function pointnetClassifierYPred = pointnetClassifier(XVal,parameters,state,isTrainingVal);% Choose prediction with highest score as the class label for% XTest.[~,YValLabel] = max(YVal,[],1);[~,YPredLabel] = max(YPred,[],1);% Collect confusion metrics.cmat = aggreateConfusionMetric(cmat,YValLabel,YPredLabel);figure;chart = confusionchart(cmat,classes);acc = sum(diag(cmat))./sum(cmat,"all")1.2.3.4.5.6.7.8.9.10.11.12.13.14.15.16.17.18.19.20.21.22.23.24.25.26.27.28.29.30.31.32.33.34.35.36.37.38.39.40.41.42.43.44.45.46.47.48.49.50.51.52.53.54.55.56.57.58.59.60.61.62.63.64.65.66.67.68.69.70.71.72.73.74.75.76.77.78.79.80.81.82.83.84.85.86.87.88.89.90.91.92.93.94.95.96.97.98.99.100.101.102.103.104.105.106.107.108.109.110.111.112.113.114.115.116.117.118.119.120.121.122.123.124.125.126.127.128.129.130.131.132.133.134.135.136.137.138.139.140.141.142.143.144.145.146.147.148.149.150.151.152.153.154.155.156.157.158.159.160.161.162.163.164.165.166.167.168.169.170.171.172.173.174.175.176.177.178.179.180.181.182.183.184.185.186.187.188.189.190.191.192.193.194.195.196.197.198.



免责声明:本文系网络转载或改编,未找到原创作者,版权归原作者所有。如涉及版权,请联系删